Protein Modeling
We have KcsA* simulation and D-Ala simulation
KcsA simulation
1. preparation
We got the PDB files (ID:5VKH) from the ncbi ,the files included the structure of the single subunit and also the complete protein make up of 4 subunits.
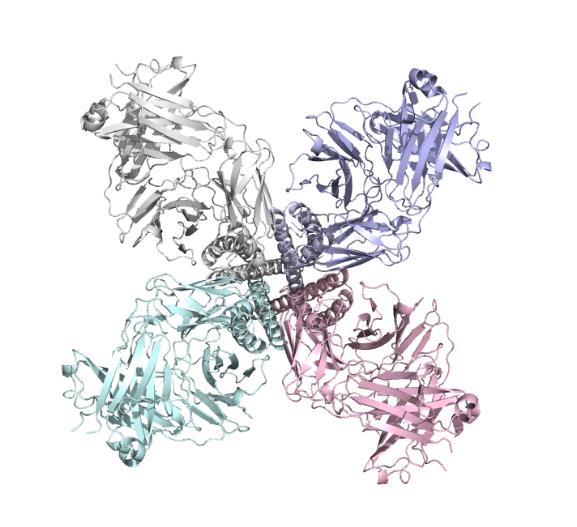
We used molecular dynamic simulation to imitate the protein in computer.The software we used is Gromacs 5.1.1
A new forcefield called GROMOSNWU is generated by adding and modifying the original GROMOS53A6 which cannot simulate a membrane.We added the parameters of lipid(dppc) from here
http://wcm.ucalgary.ca/tieleman/downloads.
2. build KcsA and KcsA*
Because the coordinate of KcsA is from the X-ray [1]diffraction of the actual protein ,we use the transmembrane domain to run the simulation as the controlled experiment.

And we change the Gly77 into D-Ala77 to build the KcsA*.


3. put the protein into membrane

First ,we orientate the symmetry axis of protein parallel to Z-axis.
Then we merge the file with another file which has a membrane from here.The membrane settles in the XY plane and makes up of 64 dppc.
4. shrink the membrane

We can see that the distance between 2 dppc is too far,which in practice must be very close to become a membrane.We need to shrink the membrane to avoid the water get in to the split of dppc-dppc and dppc-protein.




 ">
">We add water into the box .We need to check whether there are some water get into the membrane and then we should remove them by manul.
We also need to balance the charge by adding ions.If there is a non-integer charge ,we use our dummy-ion here.

6. run MD
Before the total MD simulation, we need to minimize the system and balance it in NVT and NPT.
7. analysis the structure We also meet some trouble.



Before-blue
after-green
8. electrophysiology simulation

protein modeing
It is similar with KcsA* simulation and much more easier

[1]Cuello LG, Cortes DM, Perozo E.The gating cycle of a K+ channel at atomic resolution.Elife. 2017 Nov.

